TOR signaling and autophagy in microalgae
José L. Crespo and M. Esther Pérez-PérezThe main goal of our research is to understand how microalgae adapt their growth and metabolism to stress and challenging environmental conditions, using the model microalga Chlamydomonas reinhardtii and the newly identified extremophilic microalga Chlamydomonas urium. To this aim, we are focused in two research lines:
- Dissecting the TOR signaling pathway in response to different nutritional stimuli. PI: José L. Crespo.
- Unraveling the molecular mechanisms underlying the redox control of autophagy in response to stress. PI: M. Esther Pérez-Pérez.
TOR (Target Of Rapamycin) is a master regulator of cell growth in all eukaryotes that integrates nutritional and energy cues. Our previous studies have demonstrated that the TOR signaling pathway is structurally and functionally conserved in C. reinhardtii (reviewed in Mallén-Poce et al. J Exp Bot 2022), and regulates fundamental processes like translation (Díaz-Troya et al. Plant Physiol 2011) and autophagy (Pérez-Pérez et al. Plant Physiol 2010). We have also shown that TOR activity responds to essential nutrients such as nitrogen, phosphorus and carbon. Our results indicated that phosphorus regulates TOR signaling via LST8, a conserved TOR complex 1 (TORC1) subunit that interacts with the kinase domain of TOR. Phosphorus starvation leads to a sharp decrease of LST8 protein abundance, which in turn results in downregulation of TOR activity (Couso et al. Plant Cell 2020). Moreover, we have recently demonstrated that inorganic carbon is a central regulator of TOR. We found that the photosynthetic assimilation of CO2 efficiently activates TOR signaling through the synthesis of key amino acids, particularly Gln (Mallén-Ponce et al. PNAS 2022). We are currently identifying the specific metabolites linking CO2 availability to TOR in response to light. In addition, we are investigating the role of TOR in chloroplast biogenesis in Chlamydomonas.
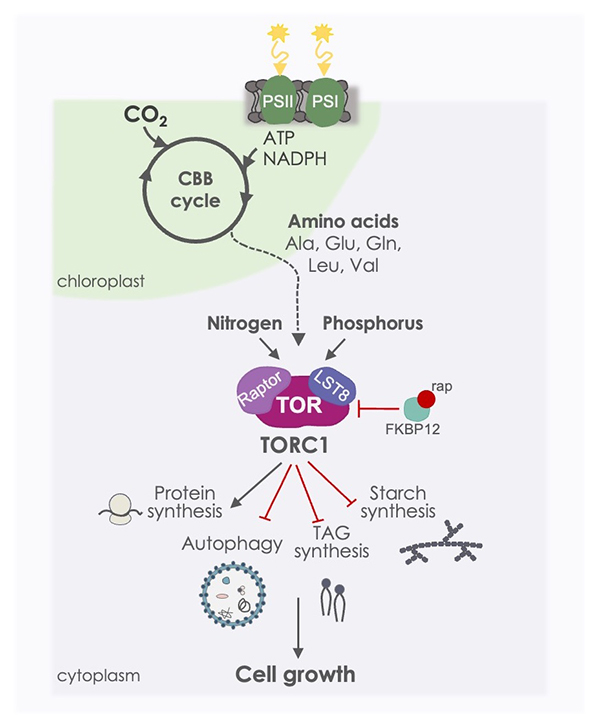
Figure 1. The TOR signaling pathway in Chlamydomonas. TORC1 is a central regulator of cell growth in Chlamydomonas by promoting anabolic processes such as protein synthesis and repressing catabolic processes including autophagy. In microalgae, TOR also controls the synthesis of lipids and starch, the two major carbon reservoirs. The TOR pathway is inhibited by the FKBP12-rapamycin (rap) complex, and responds to nutrients such as nitrogen, phosphorus and CO2.
Autophagy is a highly conserved degradative mechanism by which eukaryotic cells eliminate and recycle internal material to maintain cellular homeostasis and cope with stress. In this catabolic pathway, damaged or toxic cellular components are engulfed in double-membrane vesicles called autophagosomes, which are delivered to the vacuole for degradation. We have been pioneers in the study of this catabolic process in microalgae. We have shown that autophagy is repressed by the TOR signaling pathway and upregulated in response to a wide range of stress conditions including nutrient limitation, metal toxicity, chloroplast damage or oxidative stress (reviewed in Mallén-Ponce et al. Free Radic Bio Med 2023). We found a strong connection between ROS production and autophagy activation in most of these stress conditions (Pérez-Pérez et al. Plant Physiol 2012). Our results demonstrate that the ATG8 lipidation system, which plays a fundamental role in autophagosome formation, is targeted by ROS. We unraveled the molecular mechanism responsible for the ROS-linked reversible inhibition of the cys-protease ATG4 (Pérez-Pérez et al. Autophagy 2014; Pérez-Pérez et al. Plant Physiol 2016). More recently, we have shown that the activity of the E2-activating enzyme ATG3 is also controlled by ROS to ensure ATG8 lipidation and autophagy progression under oxidative stress (Mallén-Ponce et al. Plant Physiol 2024). We are currently investigating the role of autophagy and ROS production in the control of carbon metabolism in Chlamydomonas cells under different nutritional and light regimes.
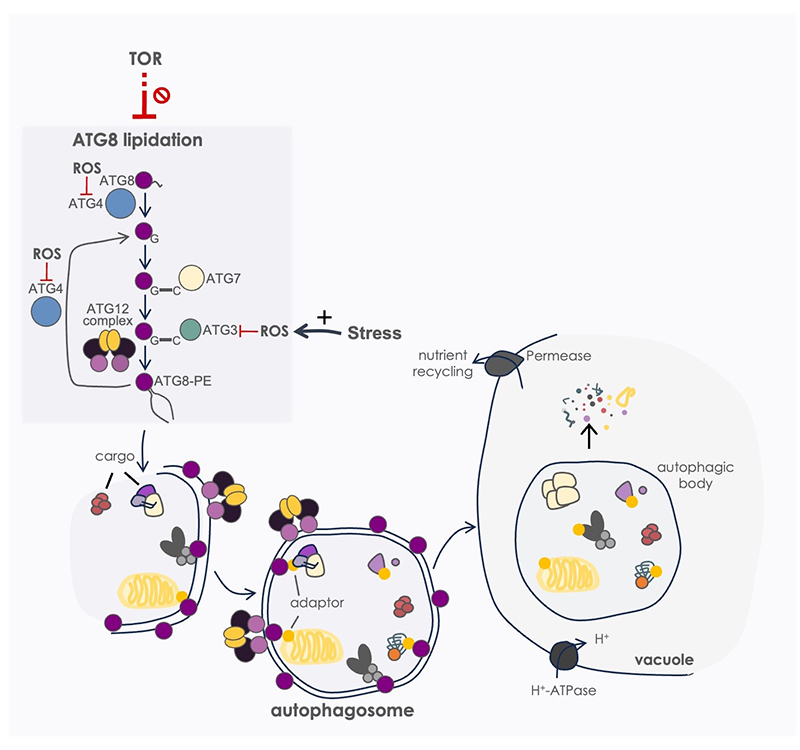
Figure 2. Autophagy in Chlamydomonas. The components of the ATG8 system (ATG3, ATG4, ATG7 and ATG8) act coordinately to promote autophagosome biogenesis. The conjugation of ATG8 to the membrane phospholipid phosphatidylethanolamine (PE) is a key step in both autophagosome formation and cargo selection. For ATG8 lipidation, first, nascent ATG8 is cleaved at a highly conserved Gly at its C-terminus by the Cys-protease ATG4. Then, by the sequential action of ATG7 and ATG3, ATG8 is bound to the headgroup of PE, resulting in the formation of the ATG8-PE adduct. The ATG5-ATG12-ATG16 complex potentiates the binding of ATG8 to PE. Besides processing ATG8, ATG4 has a delipidating activity, cleaving ATG8-PE and releasing free ATG8 from autophagosome membranes. The autophagosome content is finally degraded and recycled in the vacuole. Autophagy is negatively regulated by TOR and activated by ROS-linked stress. Two main autophagy regulatory proteins, ATG4 and ATG3, are reversibly inhibited by ROS.
Our group has also initiated a new research line focused on the study of TOR signaling and autophagy in extremophilic microalgae, since these organisms have to adapt their growth to environmental extreme conditions. To this aim, we have isolated a new extremophilic microalga, Chlamydomonas urium, from the water source of the Tinto river (Nerva), an extremely acidic river located in the South of Spain. We have reported that C. urium exhibits a high photosynthetic capacity as well as an active recycling of lipid droplets through a selective autophagy pathway known as lipophagy (Pérez-Pérez et al. New Phytologist 2024).

| Name | Surname | Category | Phones | |
|---|---|---|---|---|
| José Luis | Crespo González | CSIC Scientifical Researcher | ext. 446074 | |
| Samuel | Gámez Arcas | Postdoctoral Researcher | ext. 446061 | |
| Manuel Jesús | Mallén Ponce | Postdoctoral Researcher | ext. 446069 | |
| Irene | Muñoz Palacios | Predoctoral Researcher | ||
| Yosu | Odriozola Gil | Predoctoral Researcher | ext. 446069 | |
| María Esther | Pérez Pérez | CSIC Tenured Scientist | ext. 446069 | |
| Andrea | Quintero Moreno | Laboratory Technician | ext. 446069 | |
| María del Águila | Ruiz Sola | Doctor Professor Contract | 954139176 ext. 446061 |
- Mallén-Ponce MJ, Quintero-Moreno AM, Gámez-Arcas S, Grossman AR, Pérez-Pérez ME, Crespo JL. Dihydroxyacetone phosphate generated in the chloroplast mediates the activation of TOR by CO2 and light. Science Advances. 2025 (in press).
- Pérez-Pérez ME, Mallén-Ponce MJ, Odriozola-Gil Y, Rubio A, Salas JJ, Martínez-Force E, Pérez-Pulido AJ, Crespo JL. Lipid turnover through lipophagy in the newly identified extremophilic green microalga Chlamydomonas urium. New Phytologist. 2024. Jul;243(1):284-298. doi.org/10.1111/nph.19811.
- Mallén-Ponce MJ, Pérez-Pérez ME. Redox-mediated activation of ATG3 promotes ATG8 lipidation and autophagy progression in Chlamydomonas reinhardtii. Plant Physiol. 2023 Dec 30;194(1):359-375. doi: 10.1093/plphys/kiad520.
- Mallén-Ponce MJ, Pérez-Pérez ME, Crespo JL. Photosynthetic assimilation of CO2 regulates TOR activity. Proc Natl Acad Sci U S A. 2022 Jan 11;119(2):e2115261119. doi: 10.1073/pnas.2115261119.
- Couso I, Pérez-Pérez ME, Ford MM, Martínez-Force E, Hicks LM, Umen JG, Crespo JL. Phosphorus Availability Regulates TORC1 Signaling via LST8 in Chlamydomonas. Plant Cell. 2020 Jan;32(1):69-80. doi: 10.1105/tpc.19.00179.
- Heredia-Martínez LG, Andrés-Garrido A, Martínez-Force E, Pérez-Pérez ME, Crespo JL. Chloroplast Damage Induced by the Inhibition of Fatty Acid Synthesis Triggers Autophagy in Chlamydomonas. Plant Physiol. 2018 Nov;178(3):1112-1129. doi: 10.1104/pp.18.00630.
- Pérez-Pérez ME, Lemaire SD, Crespo JL. Control of Autophagy in Chlamydomonas Is Mediated through Redox-Dependent Inactivation of the ATG4 Protease. Plant Physiol. 2016 Dec;172(4):2219-2234. doi: 10.1104/pp.16.01582.
- Pérez-Pérez ME, Zaffagnini M, Marchand CH, Crespo JL, Lemaire SD. The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy. 2014;10(11):1953-64. doi: 10.4161/auto.34396.
- Pérez-Pérez ME, Couso I, Crespo JL. Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy. 2012 Mar;8(3):376-88. doi: 10.4161/auto.18864.
- Pérez-Pérez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010 Apr;152(4):1874-88. doi: 10.1104/pp.109.152520.


