Regulatory RNAs in cyanobacteria
Alicia M. Muro-Pastor and Agustín VioqueCyanobacteria are photosynthetic organisms with a versatile metabolism, minimal nutritional requirements and a remarkable ability to adapt to changing environments. As an example, their major “nutrient”, sunlight, is subjected to daily cycles of light and darkness as well as to changes in its intensity depending on time of day and time of year. In addition, complex cyanobacteria such as Nostoc sp., our model organism, are able to carry out several cell differentiation processes, including differentiation of heterocysts (specialized cells devoted to fixation of atmospheric nitrogen in the absence of combined nitrogen).
Small non-coding RNAs (sRNAs) and antisense RNAs (asRNAs) are currently recognized as post-transcriptional regulators of virtually every aspect of bacterial physiology and essential elements of the regulatory circuits operated by most transcriptional regulators. Our current work aims at understanding the regulation of adaptation to different stress situations, from the perspective of the implication of regulatory RNAs. We are applying global approaches, including RNA-Seq (Mitschke et al, 2011; Brenes-Álvarez et al. 2023), design of algorithms for the prediction of sRNAs in cyanobacterial genomes (Brenes-Álvarez et al., 2016) or construction of co-expression networks (Brenes-Álvarez et al., 2019). These approaches have allowed us to identify a large number of sRNAs and asRNAs whose transcription is regulated in response to various environmental stresses, so they would be candidates to be involved in regulatory mechanisms related to adaptation to these stress situations.
We have studied and characterized several sRNAs whose expression is induced under nitrogen limitation conditions. Thus, for example, NsiR4 (nitrogen stress inducible RNA 4) regulates the expression of glutamine synthetase in Synechocystis (Klähn et al., 2015) and of Calvin cycle enzymes in Nostoc (Brenes-Álvarez et al., 2021), where the level of NsiR4 is much higher in heterocysts than in vegetative cells (figure).
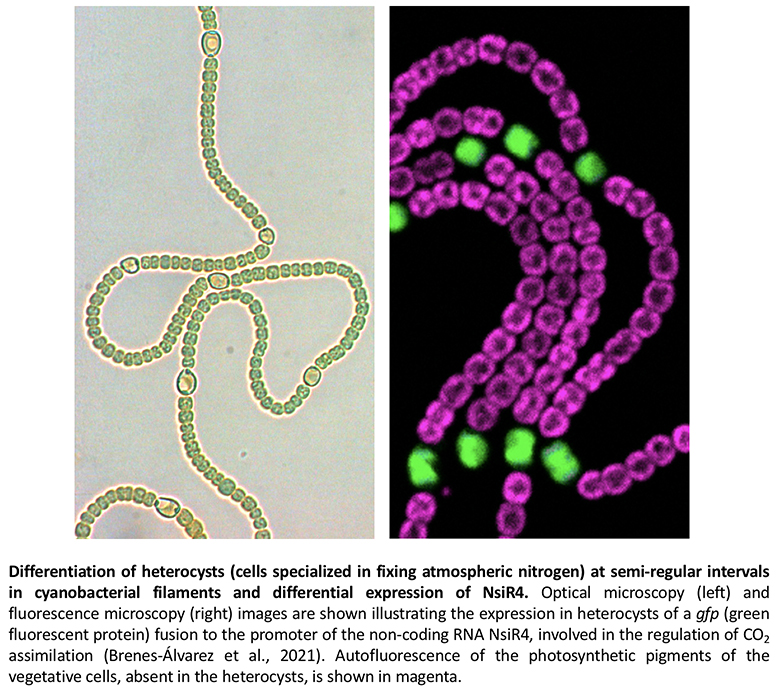
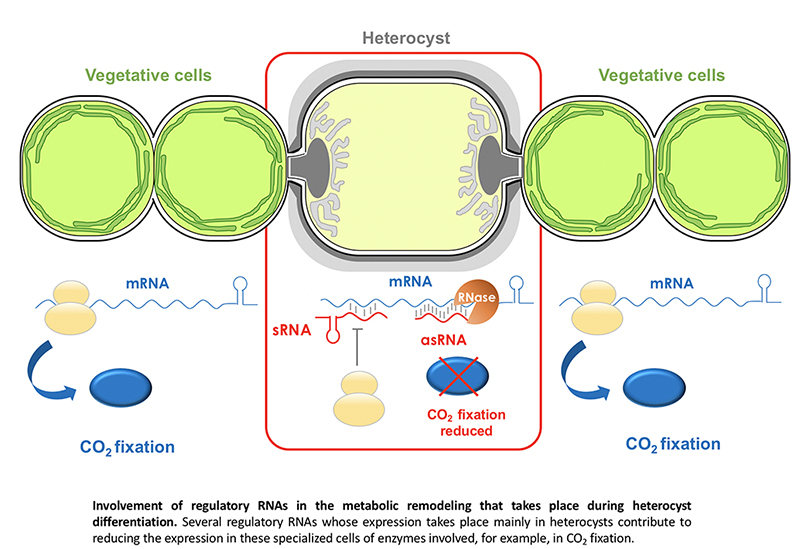
Heterocyst differentiation involves transcriptional patterns that are exclusive of these cells specialized in the fixation of atmospheric nitrogen. We have identified several sRNAs and asRNAs whose expression occurs exclusively in heterocysts. Among the sRNAs in this category is, for instance, NsiR1, transcribed very early during the differentiation process (Muro-Pastor, 2014), and which represses the expression of at least two genes related to the regulation of heterocyst development (Brenes-Álvarez et al., 2020, 2022). We have also characterized antisense RNAs that are expressed exclusively in heterocysts and regulate the expression of the glpX (fructose-1,6-bisphosphatase) or gltA (citrate synthase) genes (Olmedo-Verd et al., 2019; Brenes-Álvarez et al., 2023), contributing to the metabolic reprogramming that leads to the reduction of photosynthetic activity in heterocysts with respect to adjacent vegetative cells (figure).
Part of our studies are carried out in collaboration with the group of Wolfgang R. Hess (Genetics and Experimental Bioinformatics, University of Freiburg, Germany).

| Name | Surname | Category | Phones | |
|---|---|---|---|---|
| Manuel | Brenes Álvarez | Postdoctoral Researcher | 954139175 ext. 446060 |
|
| Sara Belén | Hernández Piñero | Postdoctoral Researcher | 954139175 ext. 446060 |
|
| Alicia María | Muro Pastor | CSIC Tenured Scientist | 954489521 ext. 446021 |
|
| Belén | Suárez Murillo | Predoctoral Researcher | 954139175 ext. 446060 |
|
| Agustín | Vioque Peña | US Professor | 954489519 ext. 446019 |
|
| Isidro | Álvarez Escribano | Postdoctoral Researcher | 954139175 ext. 446060 |
- Álvarez-Escribano I, Suárez-Murillo B, Brenes-Álvarez M, Vioque A, Muro-Pastor AM. Antisense RNA regulates glutamine synthetase in a heterocyst-forming cyanobacterium. Plant Physiol. 2024 Jul 31;195(4):2911-2920. https://doi.org/10.1093/plphys/kiae263
- Brenes-Álvarez M, Vioque A, Muro-Pastor AM. Nitrogen-regulated antisense transcription in the adaptation to nitrogen deficiency in Nostoc sp. PCC 7120. PNAS Nexus. 2023 Jun 2;2(6):pgad187. https://doi.org/10.1093/pnasnexus/pgad187
- Brenes-Álvarez M, Vioque A, Muro-Pastor AM. The heterocyst-specific small RNA NsiR1 regulates the commitment to differentiation in Nostoc. Microbiol Spectr. 2022 Apr 27;10(2):e0227421. https://journals.asm.org/doi/epub/10.1128/spectrum.02274-21
- Song K, Baumgartner D, Hagemann M, Muro-Pastor AM, Maaß S, Becher D, Hess WR. AtpΘ is an inhibitor of F0F1 ATP synthase to arrest ATP hydrolysis during low-energy conditions in cyanobacteria. Curr Biol. 2022 Jan 10;32(1):136-148.e5. https://doi.org/10.1016/j.cub.2021.10.051
- Brenes-Álvarez M, Olmedo-Verd E, Vioque A, Muro-Pastor AM. A nitrogen stress-inducible small RNA regulates CO2 fixation in Nostoc. Plant Physiol. 2021 Oct 5;187(2):787-798. https://doi.org/10.1093/plphys/kiab309
- Brenes-Álvarez M, Minguet M, Vioque A, Muro-Pastor AM. NsiR1, a small RNA with multiple copies, modulates heterocyst differentiation in the cyanobacterium Nostoc sp. PCC 7120. Environ Microbiol. 2020 Aug;22(8):3325-3338. doi: 10.1111/1462-2920.15103. Imagen de portada
- Brenes-Álvarez M, Vioque A, Muro-Pastor AM. The integrity of the cell wall and its remodeling during heterocyst differentiation are regulated by phylogenetically conserved small RNA Yfr1 in Nostoc sp. Strain PCC 7120. mBio. 2020 Jan 21;11(1):e02599-19. doi: 10.1128/mBio.02599-19
- Brenes-Álvarez M, Mitschke J, Olmedo-Verd E, Georg J, Hess WR, Vioque A, Muro-Pastor AM. Elements of the heterocyst-specific transcriptome unravelled by co-expression analysis in Nostoc sp. PCC 7120. Environ Microbiol. 2019 Jul;21(7):2544-2558. doi: 10.1111/1462-2920.14647
- Klähn S, Schaal C, Georg J, Baumgartner D, Knippen G, Hagemann M, Muro- Pastor AM, Hess WR. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci U S A. 2015 Nov 10;112(45):E6243-52. doi: 10.1073/pnas.1508412112.
- Muro-Pastor AM. The heterocyst-specific NsiR1 small RNA is an early marker of cell differentiation in cyanobacterial filaments. mBio. 2014 May 13;5(3):e01079-14. doi: 10.1128/mBio.01079-14.


